
- #INMR ADD STRUCTURE TO NMR VERIFICATION#
- #INMR ADD STRUCTURE TO NMR SOFTWARE#
- #INMR ADD STRUCTURE TO NMR DOWNLOAD#
- #INMR ADD STRUCTURE TO NMR FREE#
Our verification tools are built upon our industry-leading NMR predictors, meaning they can help you confidently confirm the identity of even your most complex structures.
#INMR ADD STRUCTURE TO NMR SOFTWARE#
Our NMR software applications fully incorporate structure in your workflow for intuitive spectral analysis: Making assignments by switching between an image of your proposed structure (hand-drawn or electronic) and your data is an inefficient and error-prone process. Our built-in verification tools help you confirm structures quickly and confidently. Our software is designed with your workflow in mind. When your spectrum/analysis is complicated though, you may lack confidence in the structure. NMR is a useful tool for synthetic chemists to quickly and nondestructively confirm the suspected identity of their sample. Spend Less Time Asking “Did I Make What I Think I Made?” Structure Identification & Verification.Whether the Der p 2 fold is a new fold or a distant relative of the α-amylase inhibitor or the immunoglobulin superfamily remains to be determined. However, a crucial element of the immunoglobulin superfamily is the Greek-key motif, and Der p 2 is lacking the strand that would define a Greek-key. Strand 4 of the immunoglobulin superfamily has been replaced with an extended loop that comprises residues 59–78 of Der p 2. An examination of other strand-loop-strand motifs in the data base revealed that the two-dimensional model of Der p 2 appears similar to that of the immunoglobulin superfamily, also shown in Fig.
#INMR ADD STRUCTURE TO NMR FREE#
The arrangement of secondary structural elements in α-amylase inhibitor is clearly unlike that of Der p 2. Photo 1: Conductor-structure contact as result of insulator assembly rotation under cross-wind loading. You may have seen benchtop and cryogen free NMR instruments from Bruker, Magritek, Nanalysis, Oxford, and Thermofisher.
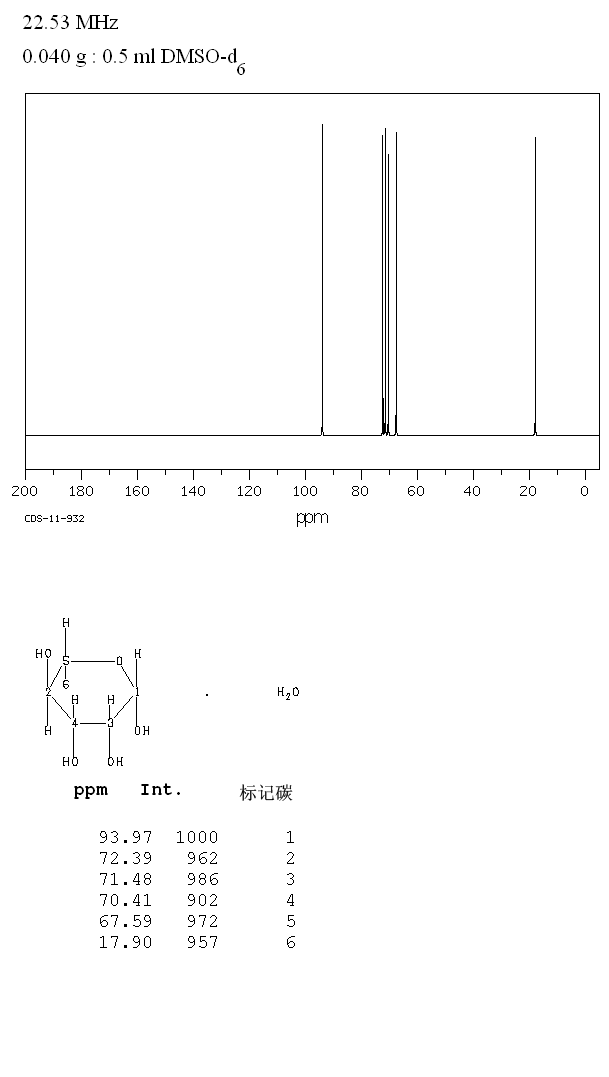
The alignment of the sheets relative to the preliminary model of Der p 2 is shown in Fig. Fluorine (19F) NMR has emerged as a useful tool for characterization of slow dynamics in 19F-labeled proteins. Only a single potential structural homologue, the α-amylase inhibitor (1hoe.pdb), was found. Since there appear to be two three-stranded anti-parallel ॆ-pleated sheets, a search of several structural data bases was initiated. A value of 1 indicates that the amide proton of that residue remained protonated for greater than 1 week after exchange into D 2O buffer.
#INMR ADD STRUCTURE TO NMR DOWNLOAD#
iNMR is a software available on Mac or Windows you can download in demo mode for use of select. Residues that could not be found in the HNHA or were too degenerate to measure accurately are indicated with a zero bar height. I need to analysis NMR data anyone has a program. These ranges are indicated by dashed lines. ॆ-sheet residues typically have coupling constants of 8–10 Hz, and α-helices typically have coupling constants of 4–7 Hz. The bar height indicates to J HN-HA coupling constant for that residue. Blanks indicate the absence of a cross-peak or a residue that was too degenerate to determine unambiguously. The bar height indicates the intensity, on an arbitrary scale, of the H α(i) − H N(i+1) NOESY cross-peak in the 15N-NOESY-HSQC, CHH-NOESY, and CN-NOESY. The bar height indicates the intensity, on an arbitrary scale, of the H N(i) − H N(i+1) NOESY cross-peak in the 15N-NOESY-HSQC. The bars indicate the predicted secondary structure for each residue based on the chemical shift of the C α, C ॆ, CO, and H α. Isolation of r Der p 2 (D1S)įigure 3 Secondary structure of rDer p 2 (D1S). Therefore, the disulfide bonds present in the final folded recombinant molecule most probably were formed during synthesis and not during the isolation procedure. Note that at no time did the extraction or refolding buffers contain any disulfide exchange reagents, e.g.mercaptoethanol, dithiothreitol, and glutathione. Solid-state NMR in particular can investigate the interactions and effects of peptides, proteins and small molecules on the structure, phase behaviour and. After dialysis, NaCl was added to 100 m mconcentration, and the sample was centrifuged (GS-34 rotor, 15,000 rpm for 20 min). The pellet was dissolved in one-fifth of the original culture volume of 6 m guanidine-HCl and dialyzed against 20 m mTris, pH 8.5, 1 m m EDTA without stirring overnight at room temperature, followed by a buffer change and an additional 4 h of dialysis. The sonicate was centrifuged (GS-34 rotor, 12,000 rpm for 20 min), and the supernatant was discarded. The cell pellet was frozen for 3–4 h at −20 ☌ and subsequently thawed and resuspended to one-twentieth of the original culture volume in TE (100 m m Tris, pH 8.5, 10 m m EDTA) and sonicated for 2 min on ice. Glycobiology and Extracellular Matrices To determine net nuclear spin (I), you simply add up the number of.


 0 kommentar(er)
0 kommentar(er)
